Unveiling Hidden Patterns in Tapeworm Evolution with Machine Learning
Unveiling Hidden Patterns in Tapeworm Evolution with Machine Learning
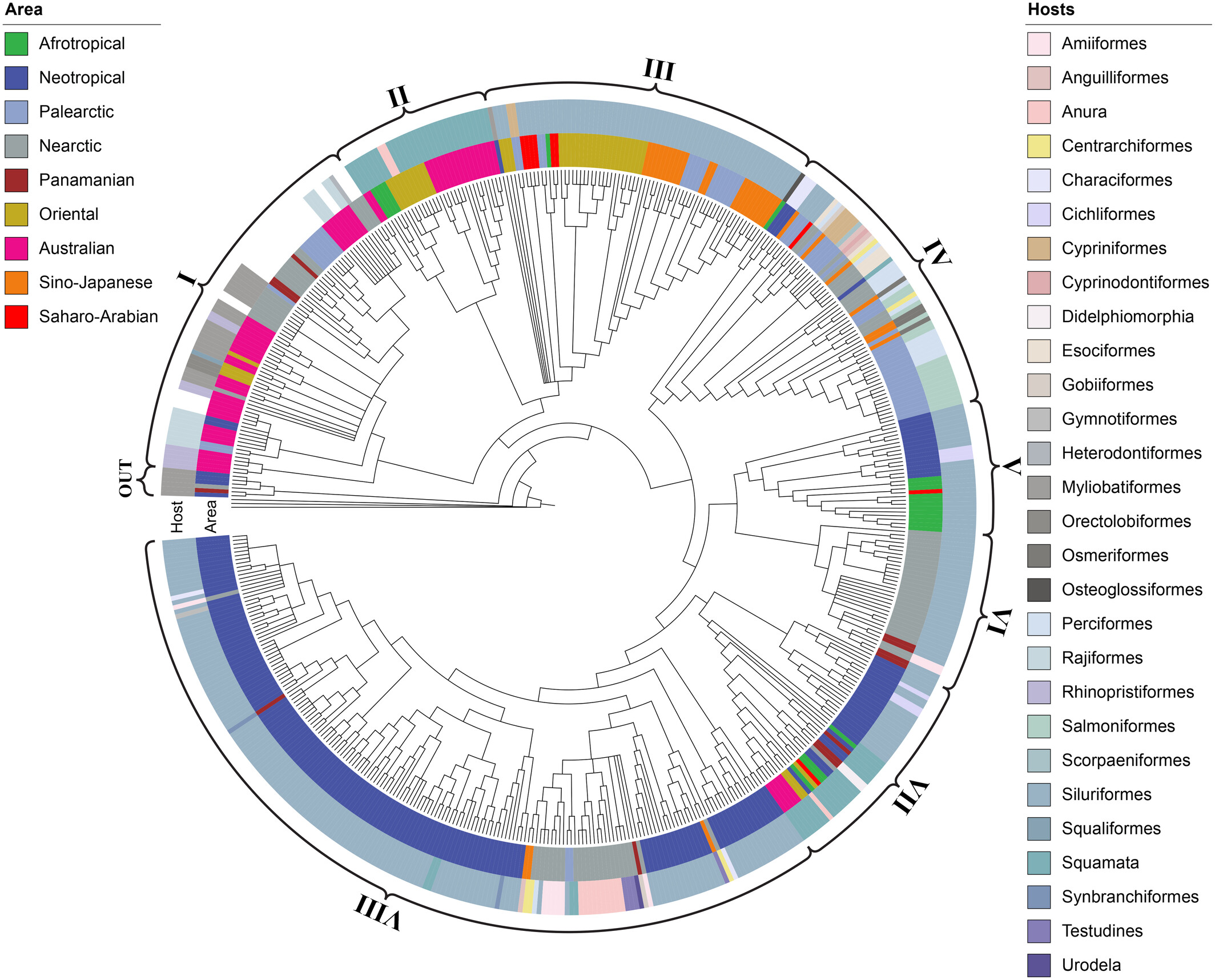
We just made a major breakthrough in understanding the evolution of proteocephalidean tapeworms using machine learning!
Our latest study, now published in Cladistics (DOI: 10.1111/cla.12610), is the first to use machine learning to uncover deep evolutionary patterns in these fascinating parasites. Traditionally, tapeworm phylogenies have been difficult to reconcile with host species and biogeography. At first glance, the evolutionary tree seems to tell one story, while host and geographic data appear disconnected. But what if there were hidden relationships beneath the surface?
Using random forests, a machine learning model, we tested whether host and biogeographical data could accurately predict the evolutionary clades of these tapeworms. The results? A striking 88.85% accuracy in classifying species into clades. This means that, despite what traditional methods to optimize host and biogeographical data could suggest, our tree does align with the expectation that host and location do play a strong role in shaping the evolution of these parasites.
This study is a game-changer. It shows that some trees that may appear, at first glance, to disagree or be disconnected from the expected host and biogeographic patterns are, in fact, more in tune with those than we can observe using traditional approaches. It also presents a new approach to unveiling hidden rules that only a multidimensional approach can reveal.
We’re now opening up a whole new way of thinking about how to correlate host and biogeography with parasite phylogenies. And you can read the entire paper for free at https://onlinelibrary.wiley.com/doi/full/10.1111/cla.12610.
Previous post
New tapeworm species named after the PI
Next post
Johannes Schultheis is now a Master!
