Phyloinformatics
We call phyloinformatics the approach that uses computational intelligence and phylogenetics to create an evolutionary framework to integrate multi-omics data to answer practical questions in One Health.
One Health is the collaborative efforts of multiple disciplines working locally, nationally, and globally to attain optimal human, animal, plant, and environmental health. Although One Health is not new, the Phyloinformatics Lab recognizes that it has become more critical in recent years. Factors that make One Health essential include the increased risk of zoonotic diseases due to contact between humans and wild animals. Additional factors are urbanization, destruction of natural habitats, climate change, and the moving of people and animals due to travel and global trade, among other factors.
In response to the continued increase in the risk of zoonoses, our lab has two main lines of research. First, investigate new pathogens’ emergence, evolution, and spread, focusing on preventing and treating infectious diseases. Second, create computational and molecular solutions to make data from biorepositories more readily available to biomedical research.
Click the link below to listen to Dr. Jacob Machado talking about the Phyloinformatics Lab at the de-CIPHERing Infectious Diseases podcast.
Support Us
Your contributions to the College of Computing & Informatics’s Bioinformatics Research Center directly empower our students and accelerate our daily research. We are deeply grateful to the private donors who help make our scientific breakthroughs possible.
Highlights
Research
Our research can be divided into four components. First, facilitating resource-efficient molecular analyses and making data from museum biorepositories more readily available to biomedical research. Second, improving genomic resources of non-model organisms with a focus on animals of particular medical or environmental interest. Third, developing phylogenetics solutions, especially if they can help improve our understanding of zoonosis. Fourth, integrating “omics” technologies (e.g., genomics, transcriptomics, metabolomics, and proteomics) to study complex host-pathogen systems in the context of One Health.
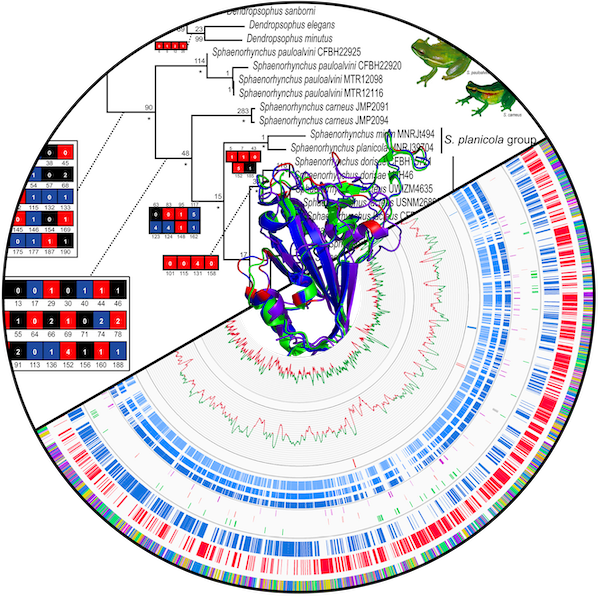
Resources
We produce bioinformatics solutions tailored to the needs of phylogeneticists and museum biorepositories. Our toolbox is diverse. For example, our genomic resources count on dedicated gene annotators for various targets, from flatworms’ mitogenomes to viruses’ genomes. Furthermore, our phylogenetic resources provide different solutions for phylogenetics result analysis, including character categorization and branch support analysis.

People
Dr. Denis Jacob Machado leads the Phyloinformatics Lab. The lab was inaugurated in August 2022 as part of UNC Charlotte’s CIPHER research center. Our collaborators include brilliant researchers from different colleges at UNC Charlotte. In addition, we also have collaborators from other institutions in the USA, Brazil, Colombia, and more. In addition, we are always happy to meet new collaborators, prospective students, and potential postdocs.

News
We stand on a tripod of research, teaching, and outreach. Whenever possible, we post about recent developments in our lab and get involved in community outreach. Keep posted about our papers, talks, posters, classes, and more on this website.
Join us
We would be thrilled if you’re considering joining the vibrant community at the Phyloinformatics Lab. We receive a fantastic number of weekly emails from individuals interested in working or collaborating with us, and your enthusiasm is genuinely appreciated!
To streamline our communication and ensure everyone gets the attention they deserve, we’ve put together a few friendly tips for reaching out, especially if you’re a student interested in job opportunities or collaboration:
- Stay updated on our latest open positions! We regularly post about them in our news section and on UNC Charlotte’s applicant site. All paid opportunities will be listed there, and we kindly ask that inquiries about such positions come in response to these posts.
- Before reaching out, take a moment to explore our lab’s mission, vision, and research areas on our homepage. Please familiarize yourself with the goals of UNC Charlotte’s CIPHER center and the Department of Bioinformatics and Genomics, our proud location. Dive into our research projects, and let us know what aspects caught your eye.
- When you contact us, share your specific interests in our lines of research and outline your overarching goals as a student or researcher. While you don’t need a fully formed research plan, we’re open to detailed ideas and suggestions for potential funding opportunities.
- Make your email uniquely yours! If it feels too generic and seems like a copy-paste message that could go to any lab, it might be mistaken for spam, and unfortunately, we won’t be able to respond.
- If your question is about applying to become a student, perhaps in one of our graduate programs in bioinformatics, you should first check the details on UNC Charlotte’s website. For example, click here to read about our Master’s in bioinformatics and click here to read about the Ph.D. program on bioinformatics and computational biology.
We’re eager to connect with individuals who share our passion for phyloinformatics, and we look forward to hearing from you. Your thoughtful approach will undoubtedly make our interaction more fruitful. Let’s embark on this exciting journey together!
